Epigenetic Gene Regulation
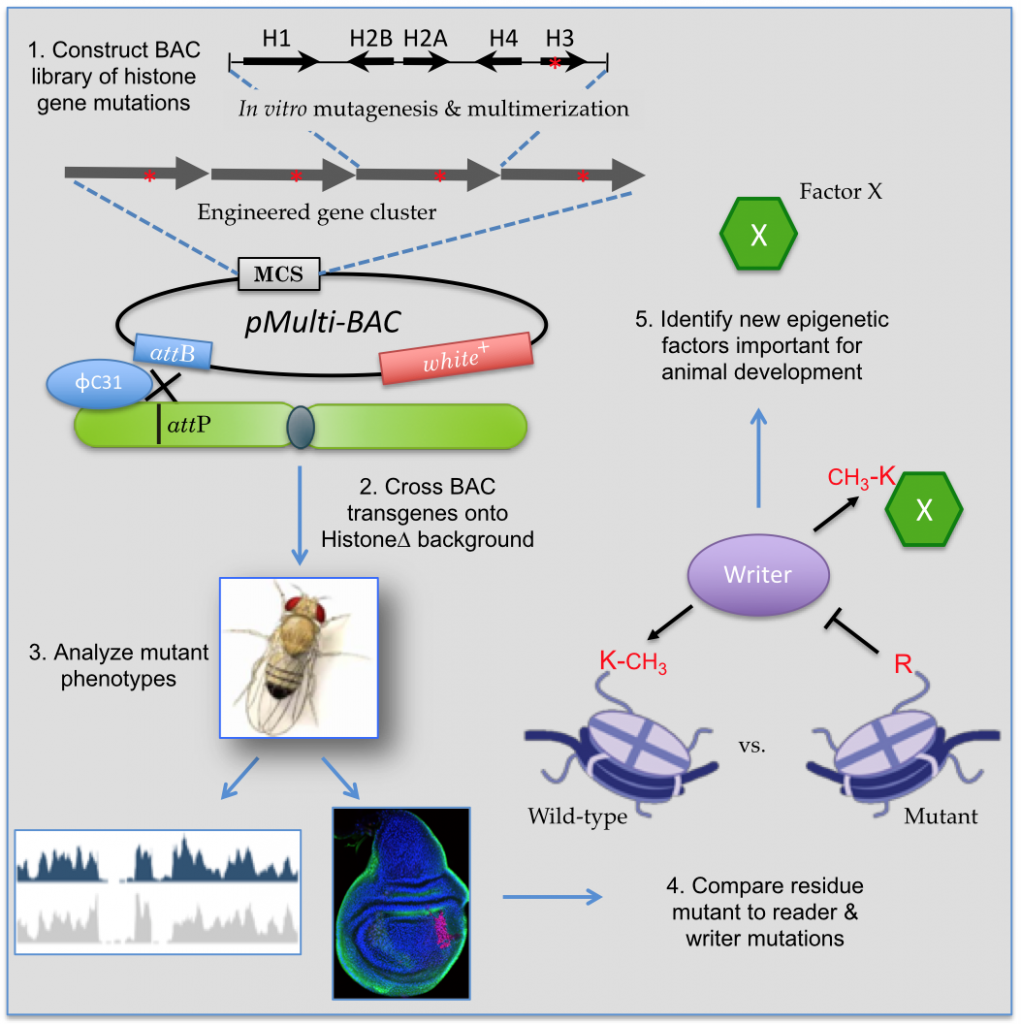
Post-translational modification (PTM) of histone and other chromatin-associated proteins is fundamental to the regulation of genome function and metazoan development. We have formed a consortium here at UNC (including the Matera, Duronio, McKay, Strahl and Marzluff labs) to create an animal model to directly test the paradigm that specific histone PTMs control epigenetic inheritance and cell fate. Because many chromatin-modifying enzymes have multiple histone or non-histone substrates, the extent to which nuclear architecture, epigenetic inheritance, cell growth and animal development is governed by particular histone PTMs is not known. Due to a lack of an animal model to study multi-copy histone genes, the biological community has been unable to determine the relative contributions that particular histone PTMs have on growth and development. Understanding this information is important, as the misregulation of histone PTMs is thought to underlie many human diseases, including cancer. Therefore, we have developed a genetic platform (see panel above) that allows manipulation of the entire histone multigene family. We engineer gene clusters to express specific histone mutants that prevent the binding of effector proteins in chromatin. These studies will thus directly evaluate the contributions of specific histone PTMs to epigenetic regulation during animal development, and will create a genetic resource for others to expand upon. Our focus is on understanding how histone PTMs modulate transcription and RNA processing reactions; open questions abound.