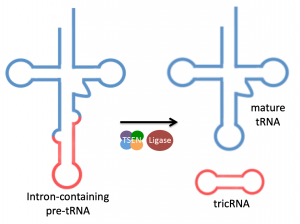
tRNA splicing and circRNAs
Hayne C.K., Butay K.J., Stewart Z.D., Krahn J.M., Pereira L., Williams J.G., Petrovitch R., Deterding L.J., Matera A.G., Borgnia M.J. and Stanley R.E. (2023). Structural basis for pre-tRNA recognition and processing by the human tRNA splicing endonuclease complex. Nature Structural and Molecular Biology 30: (Epub 23 May 2023). PMID: 37231153
Chen L-L. , Bindereif A., Bozzoni, I., Chang H-Y., Matera A.G., Gorospe. M, Hansen T.B., Kjems J., Ma X-K., Rajewsky N., Salzman J., Pek P.J., Wilusz J.E., Yang L., Zhao F. (2023). A guide to naming eukaryotic circular RNAs. Nature Cell Biology 25: 1-5. PMC10114414
Schmidt C.A., Min L., McVay M., Giusto J.D., Brown J.C., Salzler H.R. and Matera A.G.* (2022). Mutations in Drosophila tRNA processing factors cause phenotypes similar to human Pontocerebellar Hypoplasia. Biology Open 11: bio.058928.
Hayne C.K., Schmidt C.A., Haique M.I., Matera A.G. and Stanley R.E. (2020). Reconstitution of the human tRNA splicing endonuclease (TSEN) complex: insight into the regulation of pre-tRNA cleavage. Nucleic Acids Research 48:7609-7622. PMID: 32476018
Schmidt C.A. and Matera A.G. (2020). tRNA introns: Presence, processing, and purpose. Wiley Interdisciplinary Reviews RNA 11: e1583. Epub 28 Dec 2019. PMID: 31883233
Schmidt C.A., Giusto J.D. and Matera A.G. (2019). Molecular determinants of metazoan tricRNA biogenesis. Nucleic Acids Res 47: 6452-6465. PMID: 31032518.
Noto J.J., Schmidt C.A. and Matera A.G. (2017). Engineering and expressing circular RNAs via tRNA splicing. RNA Biology 14:978-984. Epub: 12 Apr 2017. doi:10.1080/15476286.2017.1317911. PMC5680671
Schmidt C.A., Noto J.J., Filonov G.S. and Matera A.G. (2016). A method for expressing and imaging abundant, stable, circular RNAs in vivo using tRNA splicing. Methods in Enzymology 572: 215-236. PMID: 27241756
Lu Z., Filonov G.S., Noto J.J., Schmidt C.A., Hatkevich T.L., Wen Y., Jaffrey S.R. and Matera A.G. (2015). Metazoan tRNA introns generate stable circular RNAs in vivo. RNA 21: 1554-1565. PMC4536317